How Induced Differentiation Media Transforms Stem Cell Research
Jan 29,2026
Induced Differentiation Media (IDM) represents a cornerstone technology in modern cell biology, regenerative medicine, and therapeutic development. Unlike standard culture media designed for cell proliferation and maintenance, IDM is a highly specialized, complex formulation engineered to actively guide stem cells or progenitor cells toward a specific, mature cell fate. This is achieved by providing a meticulously orchestrated cocktail of biochemical cues,including growth factors, cytokines, hormones, and small molecules,that recapitulate the signaling pathways governing embryonic development and tissue homeostasis in vivo.
Table of Contents
1.What is Induced Differentiation Media?
2.How Does Induced Differentiation Media Work?
3.A Comparative Analysis: Situating IDM in the Cell Engineering Toolkit
4.A Typology of Induced Differentiation Media by Target Lineage
5.Applications, Challenges, and Future Directions
01 What is Induced Differentiation Media
At its core, induced differentiation media is a purpose-built cell culture system designed not merely to sustain cells, but to transform them. It is defined as a specially formulated medium whose primary function is to actively guide cells from a less specialized state, such as a pluripotent or multipotent stem cell, to a more specialized, terminally differentiated state. This process involves the strategic addition of specific inducers that activate latent intracellular genetic programs, compelling the cell to undergo profound changes in morphology, gene expression profile, and physiological function, ultimately adopting the identity of a target cell type like a neuron, heart muscle cell, or liver cell.
The composition of IDM is intricate and context-dependent, tailored precisely to the desired differentiation outcome. However, all formulations are built upon a common structural framework.
Core Components of Induced Differentiation Media
The efficacy of any induced differentiation protocol hinges on the precise combination and concentration of its components. These can be broadly categorized into a basal medium, a suite of inducing factors, and essential supplements.
1.Basal Medium: Serves as the foundational nutrient broth (e.g., DMEM, RPMI 1640), providing essential salts, glucose, amino acids, and vitamins.
2.Inducing Factors: The “active ingredients” that direct lineage commitment. These include:
● Growth factors and cytokines (e.g., BMPs, FGFs, Activin A, VEGF)
● Small molecules (e.g., CHIR99021, SB431542, retinoic acid)
● Hormones (e.g., dexamethasone, insulin)
● Chemical inducers (e.g., IBMX, indomethacin)
3.Supplements: Such as serum-free formulations (N2, B27), non-essential amino acids, and antibiotics.
This precise chemical language enables researchers to steer stem cell differentiation along defined trajectories, mimicking the natural signals present during cell differentiation embryo development. Whether applied to iPSC differentiation, MSC differentiation, or haematopoietic stem cell differentiation, IDM provides the environmental cues necessary to generate functional differentiated cells examples including neurons, osteoblasts, adipocytes, and erythrocytes [1]. The concept builds directly on early demonstrations that adult human mesenchymal stem cells can be driven into multiple lineages through defined media conditions [2].
02 How Does Induced Differentiation Media Work
IDM functions by simulating the spatiotemporally regulated signaling environment of embryogenesis, thereby activating conserved developmental pathways that orchestrate cell differentiation and stem cells fate decisions.
Key mechanisms include:
● Wnt/β-catenin Pathway: Modulated by small molecules like CHIR99021 to drive mesoderm formation(a critical step in cardiomyocyte differentiation and differentiation of mesenchymal stem cells into bone or cartilage).
● TGF-β Superfamily: High Activin A induces definitive endoderm (precursor to hepatocytes and pancreatic β-cells), while BMP4 supports ectodermal and mesodermal fates. In contrast, inhibition via SB431542 is central to neural induction.
● FGF/MAPK and SHH Pathways: Essential for patterning during differentiation of neural stem cells, particularly in generating dopaminergic neurons for Parkinson’s disease models.
● Notch Signaling: Regulates binary fate choices in differentiation of hematopoietic stem cells, influencing whether progenitors become myeloid or lymphoid lineages.
These pathways converge on master transcription factors (e.g., PU.1 for myeloid cells, SOX17 for endoderm) that remodel chromatin and initiate lineage-specific gene expression programs. In protocols using embryoid bodies (3D aggregates of pluripotent stem cells that spontaneously differentiate) IDM refines this process by suppressing off-target lineages and enhancing homogeneity [3].
For instance, in bone marrow stem cell differentiation, cytokine cocktails (SCF, IL-3, EPO) in IDM drive hematopoietic progenitors toward erythroid, megakaryocytic, or granulocytic fates, recapitulating in vivo hematopoiesis with high fidelity [4].
Fig. 1 Mechanisms of Cell Fate Specification by Induced Differentiation Media.
03 A Comparative Analysis: Situating IDM in the Cell Engineering Toolkit
To fully appreciate the unique role of induced differentiation media, it is essential to compare it with other fundamental tools in cell culture and engineering: standard cell culture media and reprogramming factors.
3.1 Induced Differentiation Media vs. Standard Cell Culture Media
Table 1. Comparison of Standard Cell Culture Media & Induced Differentiation Media
| Feature | Standard Cell Culture Media (SCM) | Induced Differentiation Media (IDM) |
| Primary Purpose | Maintenance and Proliferation: To support basic cell survival, growth, and expansion of cell populations in an undifferentiated or stable state. | Directed Specialization: To actively induce a change in cell identity, guiding cells from a progenitor state to a specific, terminally differentiated state. |
| Composition | General and Supportive: Consists of a basal medium with essential nutrients, often supplemented with a broad-spectrum growth stimulant like fetal bovine serum (FBS) or basic growth factors to encourage proliferation. | Specific and Instructive: Consists of a basal medium plus a highly specific, defined cocktail of inducing agents (growth factors, small molecules, hormones) designed to activate particular signaling pathways and drive a specific lineage commitment. |
| Functional Outcome | Cells increase in number while retaining their original identity. The goal is quantity and viability. | Cells undergo a fundamental change in their phenotype, function, and gene expression profile. The goal is a qualitative transformation. |
| Variability | Relatively standardized formulations (e.g., DMEM, RPMI 1640) are widely used across many cell types. | Highly variable and customized. The formulation for generating a neuron is drastically different from that for a liver cell, and protocols are often multi-staged, with the media composition changing over days or weeks . |
3.2 Induced Differentiation Media vs. Reprogramming Factors
Table 2. Comparison of Induced Differentiation Media & Reprogramming Factors
| Category | Reprogramming Factors | Induced Differentiation Media (IDM) |
| Fundamental Goal | Reset cell to pluripotent state (de-differentiation) | Guide cell toward specific mature fate (differentiation) |
| Primary Mechanism | Forced expression of transcription factors (e.g., Yamanaka factors) | External signals (growth factors, small molecules) |
| Starting Cell | Differentiated somatic cells (e.g., skin fibroblasts) | Pluripotent or multipotent stem cells |
| Efficiency | Low (<1%) | High (>90% with optimized protocols) |
| Primary Use in Workflow | First step: generate iPSCs | Second step: differentiate iPSCs into target cell types |
04 A Typology of Induced Differentiation Media by Target Lineage
The vast diversity of cell types in the human body necessitates an equally diverse array of differentiation media. IDM is best classified by the specific cell lineage it is designed to produce. The following sections detail the principles and typical components for generating several key cell types for research and therapeutic applications.
4.1 Mesenchymal Lineages
Differentiation of mesenchymal stem cells (MSCs) into osteoblasts, chondrocytes, or adipocytes is a hallmark assay. Osteogenic IDM includes dexamethasone, ascorbic acid, and β-glycerophosphate; adipogenic IDM uses IBMX, insulin, and PPARγ agonists. These protocols are routinely applied to bone marrow stem cell differentiation studies.
4.2 Neural Lineages
Neural induction via dual SMAD inhibition (SB431542 + LDN-193189) efficiently generates neural rosettes from iPSCs. Subsequent patterning with SHH and FGF8 yields midbrain dopaminergic neurons(key for modeling Parkinson’s disease and studying differentiation of neural stem cells )[5].
4.3 Cardiac and Hematopoietic Lineages
Cardiomyocyte differentiation employs temporal Wnt modulation (“GiWi” protocol). Meanwhile, haematopoietic stem cell differentiation relies on cytokine gradients in IDM to produce erythrocytes, T-cells, or macrophages(enabling in vitro modeling of blood disorders).
Embryoid bodies often serve as an intermediate structure in multi-lineage differentiation, though monolayer protocols now dominate for reproducibility.
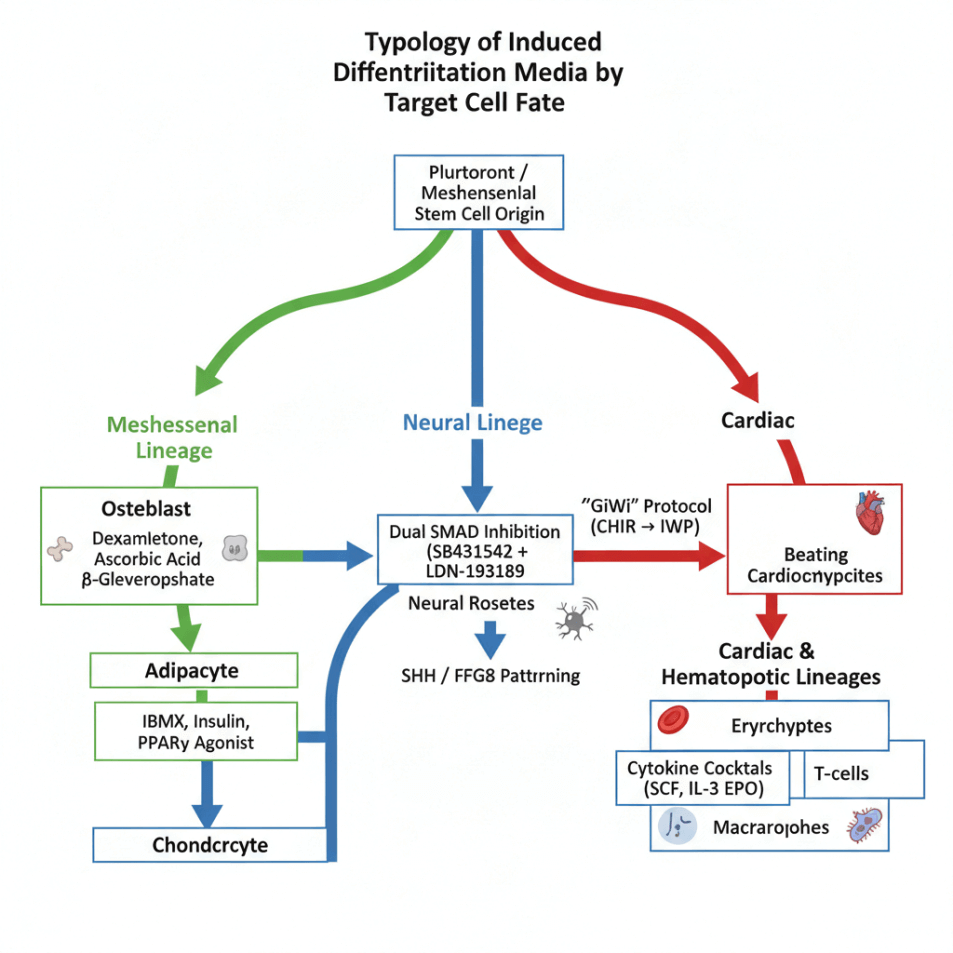
Fig. 2 Lineage-Directed Formulations of Induced Differentiation Media.
05 Applications, Challenges, and Future Directions
5.1 Core Applications
● Regenerative Medicine: Clinical trials use IDM-derived cells for macular degeneration (RPE cells), Parkinson’s (dopaminergic neurons), and heart failure (cardiomyocytes) [6].
● Disease Modeling: Patient-specific iPSC differentiation enables “disease-in-a-dish” platforms for ALS, Alzheimer’s, and leukemia [7].
● Drug Screening: IDM-generated hepatocytes and cardiomyocytes improve toxicity prediction [8].
5.2 Emerging Frontiers
Organoid-specific IDM supports 3D self-organization of brain, gut, or liver organoids, better mimicking in vivo architecture and enabling study of cell differentiation embryo-like processes [9].
5.3 Key Challenges
● Scalability & Cost: Growth factors remain expensive; shift toward small-molecule-only protocols is ongoing [10].
● Maturity & Purity: In vitro-derived cells often exhibit fetal-like phenotypes; residual undifferentiated cells risk teratoma formation [11].
● Regulatory Hurdles: GMP-compliant, xeno-free IDM is required for clinical translation under FDA/EMA oversight.
References:
[1] Keller, G. (2005). Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes & Development, 19(10), 1129–1155.
[2] Pittenger, M. F., et al. (1999). Multilineage potential of adult human mesenchymal stem cells. Science, 284(5411), 143–147.
[3] Itskovitz-Eldor, J., et al. (2000). Differentiation of human embryonic stem cells into embryoid bodies comprising the three embryonic germ layers. Molecular Medicine, 6(2), 88–95.
[4] Doulatov, S., Notta, F., Laurenti, E., & Dick, J. E. (2012). Hematopoiesis: a human perspective. Cell Stem Cell, 10(2), 120–136.
[5] Chambers, S. M., et al. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnology, 27(3), 275–280.
[6] Mandai, M., et al. (2017). Autologous induced stem-cell–derived retinal cells for macular degeneration. New England Journal of Medicine, 376(11), 1038–1046.
[7] Grskovic, M., et al. (2011). Induced pluripotent stem cells—opportunities for disease modelling and drug discovery. Nature Reviews Drug Discovery, 10(12), 915–929.
[8]Sharma, A., et al. (2017). High-throughput screening of tyrosine kinase inhibitors for cardiotoxicity using human iPSC-derived cardiomyocytes. Toxicological Sciences, 159(1), 198–207.
[9]Lancaster, M. A., & Knoblich, J. A. (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science, 345(6194), 1247125.
[10]Burridge, P. W., et al. (2014). Chemically defined generation of human cardiomyocytes. Nature Methods, 11(8), 855–860.
[11]Paull, D., et al. (2015). Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nature Methods, 12(8), 775–778.
Prev: Cardiac Endothelial Cell Isolation Keeps Failing? Copy This High-purity Playbook
Next: Targeting the Root of Cancer: A Practical Guide to Cancer Stem Cell Research


